Contents
- Cell select analysis
- load the file info to variable in workspace
- Load Parameters
- load file to variable in workspace
- scaling image from 0-255
- Display Scaled Images 1st and nth
- Create bw mask from scaled image
- Plot frames 1-20 of BWmaskclean
- If skipping the shift correction set an array Icellsx as the
- Perform Image shift Adjustment by Crosscorrelation
- Plot the Background Fluorescence Intensity (scaled with bar) and publish
- Select single/isolated cells of interest from an
- Track cells by using centroids of in the first frame
- Create an array of cell coordinates for printing cell names
- Correct the cell selection bwconncomp, error in selecting cells that
- Display each frame with cells of interest tagged by colors
- Get Restricted cell area intensities
- Compute Descriptive Statistics for each cell
- Make a color index to color cells
- Make a character string of numbers for all cells
- Make marker symbol lookup table
- Display and publish the 1st and last image (in original intensity)
- Plot Intensity vs Time for each cell and publish
- Plot Intensity vs Time(frame) and publish
- Plot a Curve Fit to response after treatment
- Save Variables
- Plot Radialwise Intensity and publish
- Save Variables
- Make Avi Movie of original images and Mean +/- SD Inensity of Cell Selection
Cell select analysis
back to top%--------------------------------------------------- %cd('C:\XXX')% where XXX = your file path to image data %---------------------------------------------------
load the file info to variable in workspace
back to top%--------------------------------------------------- %files: % 'B8_treatment_0,1_M_f-14_(GFP,0,2Hz,180f).tif' % 'C8 psychosinie 5,0 M_f-12_(GFP, 0.2Hz, 180f).tif' % 'D8 vehicle_f-12_(GFP, 0.2Hz, 180f).tif' % 'T3-A04_5uM psychosine_40_(GFP, 0.1Hz, 80).tif' %--------------------------------------------------- %Trial 30_05/11/12 %file = 'B2 f-12 treatment XuM H2O (conditions).tif'; %where B2=well number, %f-12=treatment frame, XuM=concentration of tretment, conditions=treatment conditions file = 'B2 f-12 treatment XuM H2O (conditions).tif'; %---------------------------------------------------
Load Parameters
back to top%--------------------------------------------------- %CellArea = 'WholeCell'; %final area may not equal initial area CellArea = 'Restricted'; %final area equals initial area Treatment = file(1:strfind(file,'.tif')-1); %remove .tif from file names if ~isempty(strfind(Treatment,'frame')) %find frame treat when frame notation used FrameTreat = Treatment(strfind(Treatment,'frame')+6:strfind(Treatment,'frame')+7); FrameTreat = str2double(FrameTreat); elseif ~isempty(strfind(Treatment,'f-')) %find frame treat when f- notation used FrameTreat = Treatment(strfind(Treatment,'f-')+2:strfind(Treatment,'f-')+3); FrameTreat = str2double(FrameTreat); elseif ~isempty(strfind(Treatment,'no treatment')) %find if no treatment FrameTreat = 0; else FrameTreat = 0; %assume not treated end FrameRate = ' (0.2 Hz)'; %0.2Hz = 1frame/5seconds %FrameRate = ' (0.1 Hz)'; SampleFreq = 0.2; %sample rate Hz TimeTreat = roundn(FrameTreat/SampleFreq/60,-1); %minute integer FramePostTreat = FrameTreat + 1; Printcolor = 'BW'; %color of plots and figures %Printcolor = 'Color'; %will print graphs in color AxisFont = 20; Itemp = imread(file); %read first frame width = size(Itemp,2); height = size(Itemp,1); clear Itemp; info = imfinfo(file); %load info of series frames = length(info); %frames = 30; %set frames to process less than all frames clear info; Well = strtok(file); %well on plate Passage = 'P-29'; %cell passage number MyDate = '2012-05-11'; %date of experiment %FLuoXunits = 'frame'; FluoXunits = 'minutes'; FluoYLim = [0 255]; %set the fluo y-axis limits FluoYTick = [0; 50; 100; 150; 200; 250]; %make standard fluo y-axis FluoYTickS = num2str(FluoYTick); %make standard fluo y-axis char FluoXt = frames/SampleFreq; %seconds FluoXt = FluoXt/60; %minutes FluoXdiv = floor(FluoXt/5); %integer space between 5 ticks min Min2Frame = FluoXdiv*60*SampleFreq; %space between 5 ticks frames FluoXLim = [1-5 frames+5]; %set the fluo x-axis limits FluoXTick = [1; 1*Min2Frame; 2*Min2Frame;... 3*Min2Frame; 4*Min2Frame; 5*Min2Frame]; %standard fluo x-axis FluoXTickS = num2str([0; 1*FluoXdiv; 2*FluoXdiv;... 3*FluoXdiv; 4*FluoXdiv; 5*FluoXdiv]); %standard fluo x-axis char %Focus = 'Foocus: 10x'; %focus on microscope %Focus = 'Focus: 20x'; Focus = 'Foxus: 40x'; if strcmp(Focus,'Focus: 10x') %pixel to distance conversion pixeldist = '3 Pixels ~ 2um'; elseif strcmp(Focus,'Focus: 20x') pixeldist = '3 Pixels ~ 1um'; else %strcmp(Focus, 'Focus: 40x') pixeldist = '3 Pixels ~ 0.5um'; end %-------------------------------------------------- %%Determine if need to do shift correction %--------------------------------------------------- shiftCorrection = 'Yes'; %shiftCorrection = 'No'; %---------------------------------------------------
load file to variable in workspace
back to top%--------------------------------------------------- I = ones(height,width,frames,'uint8'); %preallocate for k = 1:frames; I(:,:,k) = imread(file,'index',k); end clear width height %---------------------------------------------------
scaling image from 0-255
back to top%--------------------------------------------------- %(01/11/12)-need to create array for storing scaled images (double, uint8) % I1 = I(:,:,1); % I1sc = double(I1); %store to array % I1sc = I1sc - min(min(I1sc)); % I1sc = I1sc * 256/(max(max(I1sc))); % I1sc = uint8(I1sc); %store to array Isc = zeros(size(I),'uint8'); Iscd = double(Isc(:,:,1)); for k = 1:frames; Iscd(:,:,1) = double(I(:,:,k)) - min(min(double(I(:,:,k)))); Isc(:,:,k) = uint8(Iscd * 256 / (max(Iscd(:)))); end clear Iscd; %---------------------------------------------------
Display Scaled Images 1st and nth
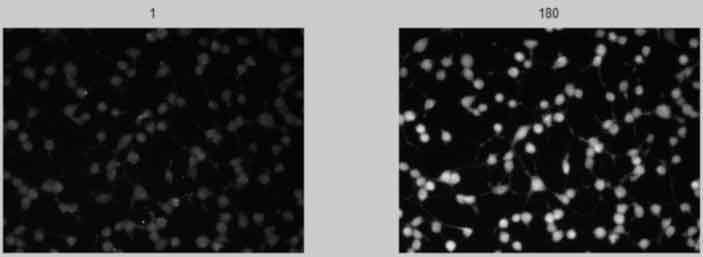
back to top
%--------------------------------------------------- % figure; % subplot(1,2,1), imshow(Isc(:,:,1)),title(num2str(1)); % subplot(1,2,2),imshow(Isc(:,:,frames)),title(num2str(frames)); %---------------------------------------------------
Create bw mask from scaled image
back to top%--------------------------------------------------- % level = graythresh(I1sc); % BWI1sc = im2bw(I1sc,level); % figure,imshow(BWI1sc) % %(01/11/12)-need to create array for storing BWIclean images % BWI1clean = bwareaopen(BWI1sc,280); % se = strel('disk',3); % BWI1clean2 = imclose(BWI1clean,se); %store to array BWIclean = logical(I); %preallocate array se = strel('disk',3); %create shifting filter disk for k = 1:frames; if k <=FrameTreat level = 0.75*graythresh(Isc(:,:,k)); %variable percent thresh else level = 0.75*graythresh(Isc(:,:,k)); %75 percent thresh end BWIsc = im2bw(Isc(:,:,k),level); BackgI(:,:,k) = uint8(I(:,:,k)).*uint8(~(BWIsc)); %show background of image BWIclean0 = bwareaopen(BWIsc,200); %delete small objects BWIclean(:,:,k) = imclose(BWIclean0,se); %get rid of pepper in objects end clear Isc level BWIsc BWIclean0 se ; %--------------------------------------------------
Plot frames 1-20 of BWmaskclean
back to top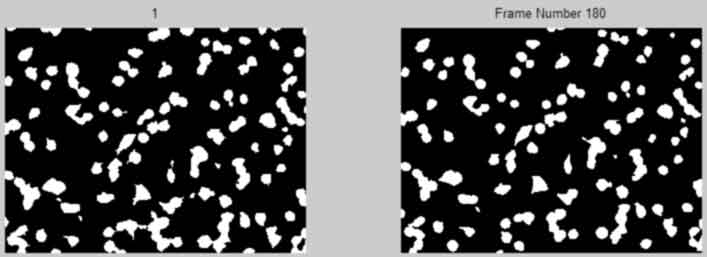
%--------------------------------------------------- % figure; % subplot(1,2,1), imshow(BWIclean(:,:,1)),title(num2str(1)); % subplot(1,2,2),imshow(BWIclean(:,:,frames)),title(num2str(frames)); % % for k = 1:20; % figure(1),imshow(BWIclean(:,:,k)) % title({['Frame Number ', num2str(k)]}); % pause(2); % end %---------------------------------------------------
If skipping the shift correction set an array Icellsx as the
back to topmask multiplied by the original images. Perform all data analysis using Icellsx, for original intensity values.
%--------------------------------------------------- if strcmp(shiftCorrection,'No') Icellsx = uint8(double(BWIclean).*double(I)); clear I BWIclean end %---------------------------------------------------
Perform Image shift Adjustment by Crosscorrelation
back to top
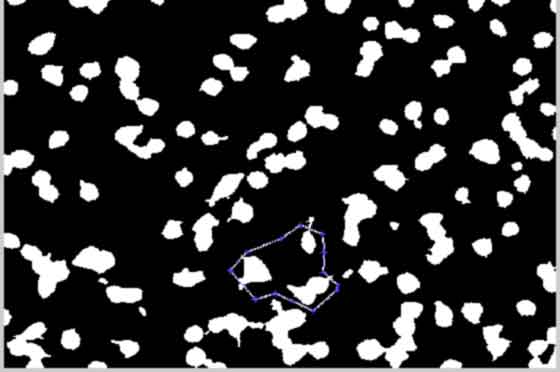
select cell(s) of interest for registering shift correction Only need to do once on first(might need to adjust later) image for stack of images
%--------------------------------------------------- if strcmp(shiftCorrection,'Yes') [BWIroiI1,xi,yi] = roipoly(double(BWIclean(:,:,1))); %return subroi xi = ceil(xi); yi = ceil(yi); roiymin = max( [ (min(yi))-125, 1 ] ); roixmin = max( [ (min(xi))-125, 1 ] ); roiymax = min( [ (max(yi))+125, size(BWIroiI1,1) ] ); roixmax = min( [ (max(xi))+125, size(BWIroiI1,2) ] ); %BWroiI1b = BWI1clean2(roiymin:roiymax, roixmin:roixmax); %larger roi A = BWIclean(:,:,1); A = A(min(yi):max(yi),min(xi):max(xi)); B = BWIroiI1(min(yi):max(yi),min(xi):max(xi)); C = A.*B; BWItemplate = C; %create and store template for shift registration clear xi yi A B C BWIroiI1 close(gcf) % figure; % imshow(BWItemplate); %-------------------------------------------------- %%Compute the offset between image 1 BWtemplate and image n %-------------------------------------------------- BWIroiI = BWIclean(uint16(roiymin:roiymax), uint16(roixmin:roixmax),:); ypeak = ones(1,frames); xpeak = ypeak; corr_offset = ones(1,2,frames); offset = corr_offset; for k = 1:frames; % BWroitemp = BWIclean2(:,:,k); % BWroiI(:,:,k) = BWroitemp(roiymin:roiymax, roixmin:roixmax); %larger roi cc = normxcorr2(BWItemplate, double(BWIroiI(:,:,k))); [max_cc, imax] = max(abs(cc(:))); [ypeak(k), xpeak(k)] = ind2sub(size(cc),imax(1)); corr_offset(1,:,k) = [ypeak(k)-size(BWItemplate,1),... xpeak(k)-size(BWItemplate,2)]; %Calculate the offset with respect to first image offset(1,:,k) = corr_offset(1,:,k) - corr_offset(1,:,1); %store to array end clear BWIroiI BWItemplate xpeak ypeak corr_offset max_cc imax clear roiymin roiymax roixmin roixmax %-------------------------------------------------- %%Offset cropping for all images with respect to image one %-------------------------------------------------- upy = min(offset(1,1,:));%max shift up (-y) from 2D image shift correlation dny = max(offset(1,1,:));%get max shift down (+y) from 2D image shift correlation lftx = min(offset(1,2,:));%get max shift left (-x) from 2D image shift correlation rgtx = max(offset(1,2,:));%get max shift right (+x) from 2D image shift correlation if upy < 0 %trim the top of canvas yminC = 1 - upy; else %no trim yminC = 1; end if dny > 0 %trim the bottom of canvas ymaxC = size(I(:,:,1),1) - dny; else %no trim ymaxC = size(I(:,:,1),1); end if lftx < 0 %trim the left of canvas xminC = 1 - lftx; else %no trim xminC = 1; end if rgtx > 0 %trim the right of canvas xmaxC = size(I(:,:,1),2) - rgtx; else %no trim xmaxC = size(I(:,:,1),2); end %-------------------------------------------------- % %shift correction for image n versus image 1 %-------------------------------------------------- for k = 1:frames; Isctemp = uint8(double(BWIclean(:,:,k)).*double(I(:,:,k))); offset2 = offset(1,:,k); Icellsx(:,:,k) = Isctemp( (yminC:ymaxC)+offset2(1),... (xminC:xmaxC)+offset2(2)); end clear cc Isctemp offset offset1 offset2 dny upy lftx rgtx %need to limit to +1 and max length or width clear I BWIclean; end %---------------------------------------------------
Plot the Background Fluorescence Intensity (scaled with bar) and publish
back to top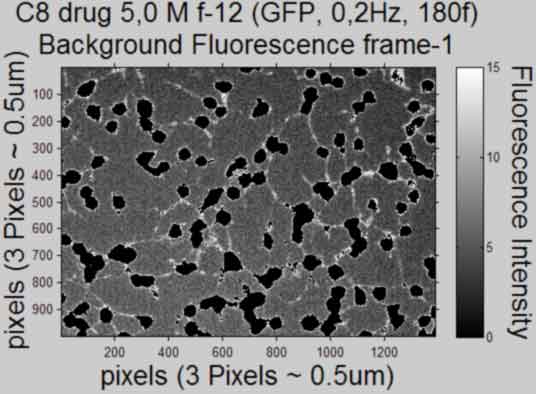
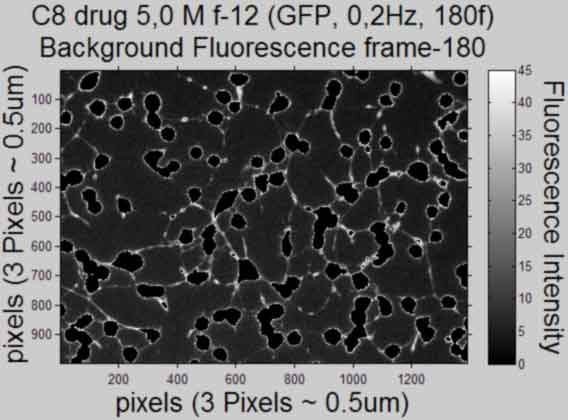
%--------------------------------------------------- for k = [1,frames] figure; if strcmp(shiftCorrection,'Yes') imshow(BackgI(yminC:ymaxC,xminC:xmaxC,k),'DisplayRange',... [min(min((BackgI(:,:,k)))),max(max((BackgI(:,:,k))))],... 'InitialMagnification', 25,'Border', 'loose'), axis on %do not scale 8bit map else imshow(BackgI(:,:,k), 'DisplayRange',... [min(min((BackgI(:,:,k)))),max(max((BackgI(:,:,k))))],... 'InitialMagnification', 25, 'Border', 'loose'), axis on %do not scale 8bit map end %imagesc(BackgI(yminC:ymaxC,xminC:xmaxC,k)), colormap gray %scaled 8bit %map set(gca,'FontSize',10) title({Treatment;... ['Background Fluorescence frame','-',num2str(k)]},'FontSize',AxisFont) xlabel(['pixels',' (',pixeldist,')'],'FontSize',AxisFont) ylabel(['pixels',' (',pixeldist,')'],'FontSize',AxisFont,... 'VerticalAlignment','baseline') hcfig = colorbar; %create colorbar and get handle set(hcfig,'FontSize', 10) cTYfig = 'Fluorescence Intensity'; hcTYfig = get(hcfig,'YLabel'); %get colorbar Yaxis title handle set(hcTYfig,'String',cTYfig,'Rotation',270,... 'VerticalAlignment','bottom','FontSize',AxisFont) %insert string %save figure saveas(gcf, [Treatment, ' Background', ' Frame ', num2str(k),... '.pdf'], 'pdf'); close gcf end clear yminC ymaxC xminC xmaxC BackgI %---------------------------------------------------
Select single/isolated cells of interest from an
back to top
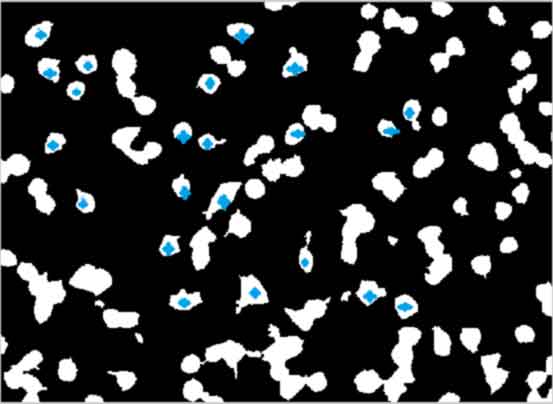
image mask of original image, for analysis
%--------------------------------------------------- k=1; BW2(:,:,k) = bwselect(double(Icellsx(:,:,k))); cc = bwconncomp(BW2(:,:,k)); STATS = regionprops(cc,(Icellsx(:,:,k)), 'BoundingBox',... 'Centroid', 'Extrema', 'PixelList', 'PixelValues','Image'); %%centroid = center of cell mass %%extrema = coordinates of cell corners %%pixelvalues = pixel intensities if strcmp(Printcolor,'Color') L(:,:,k) = labelmatrix(cc); RGB(:,:,:,k) = label2rgb(L(:,:,k)); end objects = cc.NumObjects; %RGB(:,:,:,k) = label2rgb(L(:,:,k),'Lines','k'); %alternate colormap close(gcf) %---------------------------------------------------
Track cells by using centroids of in the first frame
back to topFind same cells in all frames. Assuming the cells are not moving/migrating more than a half length of a cell beween sequential images
%--------------------------------------------------- try %attempt to find cell with given centroid else change centroid for k = 2:frames centroids = cat(1,STATS(:,k-1).Centroid); %use k-1 centroid to find %kth cell %centroids = cat(1,STATS(:,1).Centroid); %use 1st centroid to find kth cell r = centroids(:,2); %y c = centroids(:,1); %x BW2 = bwselect(Icellsx(:,:,k),c,r); cc = bwconncomp(BW2); STATS(:,k) = regionprops(cc, (Icellsx(:,:,k)),... 'BoundingBox', 'Centroid', 'Extrema',... 'PixelList', 'PixelValues','Image'); if strcmp(Printcolor,'Color') L(:,:,k) = labelmatrix(cc); RGB(:,:,:,k) = label2rgb(L(:,:,k)); %RGB(:,:,:,k) = label2rgb(L(:,:,k),'Lines', 'k'); end end clear BW2 centroids r c cc catch err if strcmp(err.identifier,'MATLAB:badsubscript') try for k = 2:frames %centroids = cat(1,STATS(:,k-1).Centroid); %use k-1 centroid to find %kth cell centroids = cat(1,STATS(:,1).Centroid); %use 1st centroid to find kth cell r = centroids(:,2); %y c = centroids(:,1); %x BW2 = bwselect(Icellsx(:,:,k),c,r); cc = bwconncomp(BW2); STATS(:,k) = regionprops(cc, (Icellsx(:,:,k)),... 'BoundingBox', 'Centroid', 'Extrema',... 'PixelList', 'PixelValues','Image'); if strcmp(Printcolor,'Color') L(:,:,k) = labelmatrix(cc); RGB(:,:,:,k) = label2rgb(L(:,:,k)); %RGB(:,:,:,k) = label2rgb(L(:,:,k),'Lines', 'k'); end end clear BW2 centroids r c cc catch err2 rethrow(err) end else rethrow(err); %if any other error stop end end %if error mismatch index dimension compare k-1 and k-2 %---------------------------------------------------
Correct the cell selection bwconncomp, error in selecting cells that
back to topshift between consecutive frames 07-5-12
%--------------------------------------------------- SwapObjects = zeros(objects,1); %initialize the swap object array for i=1:objects; cent1 = STATS(i,1).Centroid; %get the first frame selection centroid n=1; for k=1:frames; cent2 = STATS(i,k).Centroid; d = sqrt((cent1(1)-cent2(1))^2 + (cent1(2)-cent2(2))^2); if d > 45 %arbitrary threshold if exceeds then centroid switched SwapObjects(i,n) = k; %store error condition frames for objects n = n+1; end end end %%Swap the flagged Objects SwapObjects_1 = ceil(mean(SwapObjects,2)/frames); %make binary array of flaged objects if sum(SwapObjects_1) == 2; %case for when only two objects are swapping n = 1; for k = 1:length(SwapObjects_1) %step through binary array if SwapObjects_1(k) == 1 TwoObjects(n) = k; %get the object indices n = n+1; end end for k = SwapObjects(TwoObjects(1),:); TwoObjects_1 = STATS(TwoObjects(1),k); %temp store STATS object 1 TwoObjects_2 = STATS(TwoObjects(2),k); %temp store STATS object 2 STATS(TwoObjects(1),k) = TwoObjects_2; %switch STATS object 1 STATS(TwoObjects(2),k) = TwoObjects_1; %switch STATS object 2 end else if sum(SwapObjects_1) > 2; %case for when 2+ objects are switching for k = 1:size(SwapObjects,2) %step through the columns for i = 1:size(SwapObjects,1) %step through the rows if SwapObjects(i,k) > 0 cent1 = STATS(i,1).Centroid; %get the objects 1st centroid %d_temp(1) = sqrt( ( n=1; for j = 1:objects; cent2 = STATS(j,SwapObjects(i,k)).Centroid; %variable, flagged frame object D(n) = sqrt( (cent1(1)-cent2(1))^2 + (cent1(2)-cent2(2))^2 ); n = n+1; end [~,minObject] = min(D); %get object index Objects_1 = STATS(i,SwapObjects(i,k)); %temp stats flagged object Objects_2 = STATS(minObject,SwapObjects(i,k)); %temp stats minD object STATS(i,SwapObjects(i,k)) = Objects_2; %switch flagged object with minD object STATS(minObject,SwapObjects(i,k)) = Objects_1; %switch minD object with flagged object end end end end end %---------------------------------------------------
Create an array of cell coordinates for printing cell names
back to topnext to cells (Extrema(2,:) = [x y] of top right corner)
%--------------------------------------------------- % pos = ones(objects,2,frames); % for k = 1:frames % for i = 1:objects % pos(i,:,k) = STATS(i,k).Extrema(2,:); %pos[x y] % end % end %---------------------------------------------------
Display each frame with cells of interest tagged by colors
back to top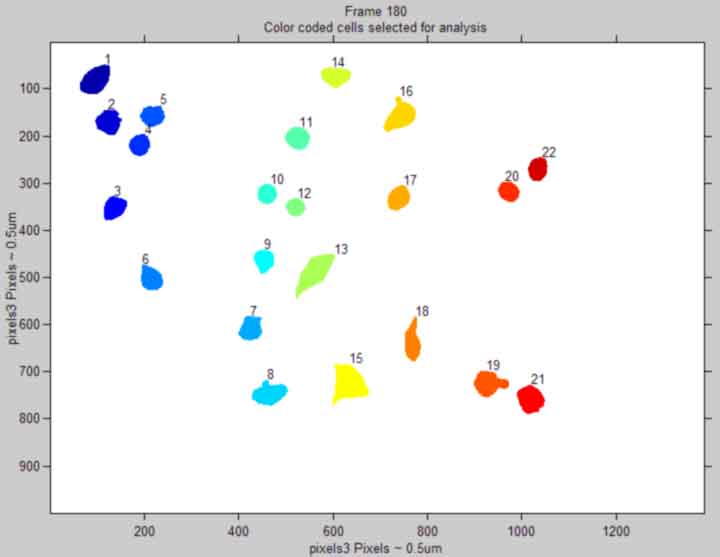
%--------------------------------------------------- % for k = 1:1 % imshow(RGB(:,:,:,k),'InitialMagnification', 25,... % 'Border', 'loose'),title({['Frame ', num2str(k)];... % 'Color coded cells selected for analysis'}) % axis on % xlabel(['pixels',pixeldist]) % ylabel(['pixels',pixeldist],'VerticalAlignment','middle') % for i = 1:objects % text(floor(pos(i,1,k)), ceil(pos(i,2,k)), num2str(i),... % 'HorizontalAlignment', 'left',... % 'VerticalAlignment', 'baseline'); % end % pause(0.3) % end % close(gcf) %---------------------------------------------------
Get Restricted cell area intensities
back to top%attempt to correct for inequal cell cropping as image contrast changes %--------------------------------------------------- for i = 1:objects for k = 1:frames Areas(i,k) = length(STATS(i,k).PixelList); %store object areas at each frame end [Sarea(i) PosSa(i)] = min(Areas(i,:)); %store smallest area size and frame for each object end Template = zeros(700); for i = 1:objects Ismall = STATS(i,PosSa(i)).Image; %smallest object image uLsmall = floor(STATS(i,PosSa(i)).BoundingBox(1:2)); %smallest image uL corner for k = 1:frames uLcurrent = floor(STATS(i,k).BoundingBox(1:2)); %current uL corner Ishift = uLsmall -uLcurrent; %determine if any difference in position of uL corners Icurrent = STATS(i,k).Image; %current object image IcurrentTemplate = Template; IcurrentTemplate(300: 300 +size(Icurrent,1) -1,... 300: 300 +size(Icurrent,2) -1) = Icurrent; %place the current object image on template IsmallTemplate = Template; IsmallTemplate(300 +Ishift: 300 +(size(Ismall,1) -1 +Ishift),... 300 +Ishift: 300 + (size(Ismall,2) -1 +Ishift)) = Ismall; Ioverlay = IcurrentTemplate.*IsmallTemplate; %compute the logical operation && Overlayidx = find(Ioverlay == 1); %find linear indices of active pixels [Irow Icol] = ind2sub(size(Ioverlay),Overlayidx); %convert linear indeces to image coordinates Irow = Irow -(300 -1); %correct for the offset placement onto the template Icol = Icol -(300 -1); %correct for the offset placement onto the template Irow = Irow +uLcurrent(2); %correct for the offset of object on original image Icol = Icol +uLcurrent(1); %correct for the offset of object on original image Overlayidx = sub2ind(size(Icellsx(:,:,k)),Irow,Icol); %get linear indices of original object embedded in original image Iorig = Icellsx(:,:,k); STATS(i,k).MyPixelValues = Iorig(Overlayidx); end end %---------------------------------------------------
Compute Descriptive Statistics for each cell
back to topArea Mean Intensity Max Intensity Standard Error of the Mean
%--------------------------------------------------- for k = 1:frames PixInt = []; for i = 1:objects if strcmp(CellArea,'WholeCell') pixInt = double(STATS(i,k).PixelValues); else pixInt = double(STATS(i,k).MyPixelValues); end areaInt(i,1,k) = length(pixInt); meanInt(i,1,k) = mean(pixInt); maxInt(i,1,k) = max(pixInt); semInt(i,1,k) = std(pixInt)/sqrt(length(pixInt)); PixInt = cat(1,PixInt,pixInt); end MeanInt(k) = mean(PixInt); %compute the mean intensity for frame StdInt(k) = std(PixInt); %compute the std for frame SemInt(k) = std(PixInt)/sqrt(length(PixInt)); %compute the SEM for frame end clear pixInt PixInt; % %%Find outliers, use function % if FrameTreat == 0; ObjectOut = zeros(1,objects); %fix later. Do not find outliers else [ObjectOut Alphas Taus] = myout3(meanInt,FrameTreat); end % %%Compute new MeanInt,StdInt,SemInt % if ~isempty(find(ObjectOut,1))%compute if outliers found for k = 1:frames PixInt = []; for i = 1:objects if strcmp(CellArea,'WholeCell') %use whole cell fluorescence pixInt = double(STATS(i,k).PixelValues); else pixInt = double(STATS(i,k).MyPixelValues); %use area select fluorescence end if ~ObjectOut(i) %if object out is not true concatenate the array PixInt = cat(1,PixInt,pixInt); end end MeanIntadj(k) = mean(PixInt); StdIntadj(k) = std(PixInt); SemIntadj(k) = std(PixInt)/sqrt(length(PixInt)); end clear pixInt PixInt; else MeanIntadj = ''; StdIntadj = ''; SemIntadj = ''; end %---------------------------------------------------
Make a color index to color cells
back to top%--------------------------------------------------- figure(1) colors = colormap; close(1) colorIdx = 1:objects; for k = 1:objects colorIdx(k) = floor( (k-1)*(64-1)/(objects-1) ) +1; end colorId = colors(colorIdx,:); clear colorIdx %---------------------------------------------------
Make a character string of numbers for all cells
back to top%--------------------------------------------------- cellnumd = (1:objects)'; cellnumd = num2str(cellnumd); %---------------------------------------------------
Make marker symbol lookup table
back to topcan only use markers if #objects <= 13
%--------------------------------------------------- mymark = ['s','o','+','d','^','p','v','*','>','x','<','.','h']; %---------------------------------------------------
Display and publish the 1st and last image (in original intensity)
back to top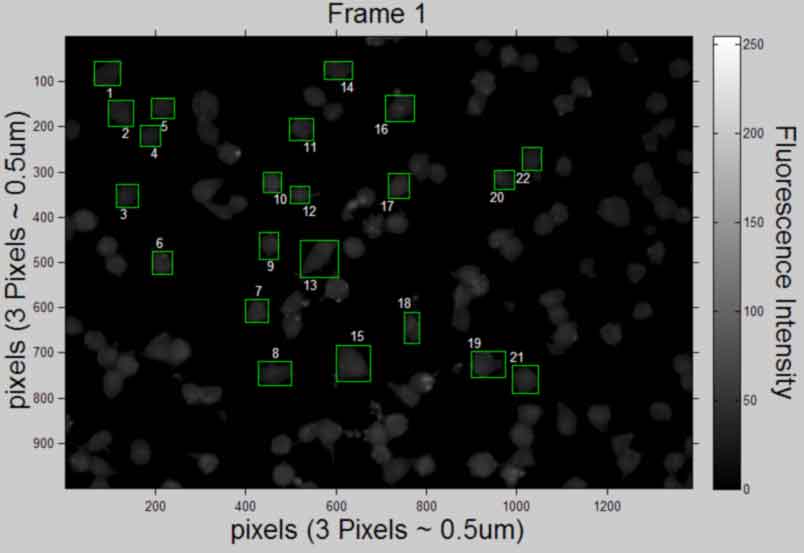
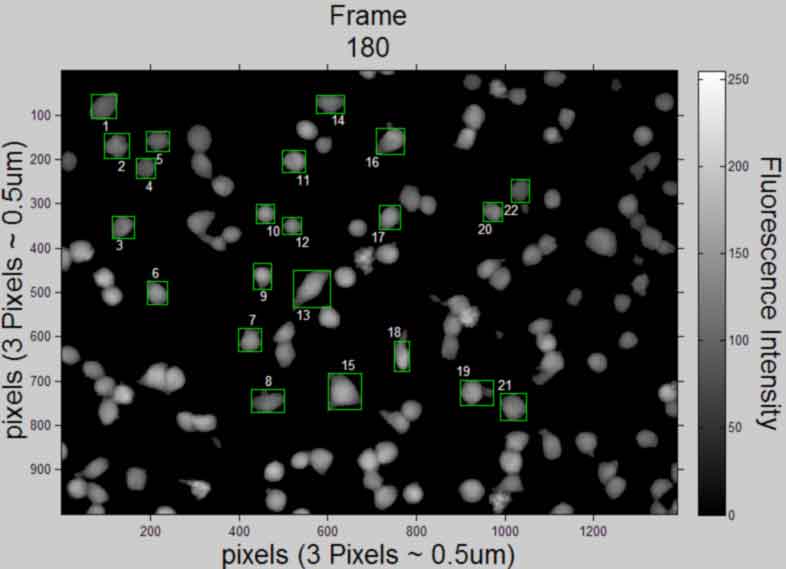
display the 1st and last image selection (in original intensity) need to store the 1st and last images I for displaying
%--------------------------------------------------- cTYfig = 'Fluorescence Intensity'; %create text % Icellsxx = uint8(logical(L).*Icellsx); % figure,imshow(Icellsxx(:,:,1),'InitialMagnification', 25,... % 'Border', 'loose'), colorbar ,title('Frame 1 selection') % xlabel('pixels') % ylabel('pixels','VerticalAlignment','middle') % hcfig = colorbar; %create colorbar and get handle % hcTYfig = get(hcfig,'YLabel'); %get colorbar Yaxis title handle % set(hcTYfig,'String',cTYfig,'Rotation',270,... % 'VerticalAlignment','bottom') %insert string % axis on; % figure,imshow(Icellsxx(:,:,frames),'InitialMagnification', 25,... % 'Border', 'loose'), colorbar ,title({'Frame', num2str(frames), 'selection'}) % xlabel('pixels') % ylabel('pixels','VerticalAlignment','middle') % hcfig = colorbar; %create colorbar and get handle % hcTYfig = get(hcfig,'YLabel'); %get colorbar Yaxis title handle % set(hcTYfig,'String',cTYfig,'Rotation',270,... % 'VerticalAlignment','bottom') %insert string % axis on; figure,imshow(uint8(Icellsx(:,:,1)),'InitialMagnification', 25,... 'Border', 'loose'), colorbar, title('Frame 1','FontSize',AxisFont) set(gca,'FontSize',10) xlabel(['pixels',' (',pixeldist,')'],'FontSize',AxisFont) ylabel(['pixels',' (',pixeldist,')'],'FontSize',AxisFont,... 'VerticalAlignment','baseline') hcfig = colorbar; %create colorbar and get handle hcTYfig = get(hcfig,'YLabel'); %get colorbar Yaxis title handle set(hcTYfig,'String',cTYfig,'Rotation',270,... 'FontSize',AxisFont,'VerticalAlignment','bottom') %insert string axis on; k = 1; %set the frame index for cell number assignment Ix = size(Icellsx,2)/2; %half length of image x-axis Iy = size(Icellsx,1)/2; %half length of image y-axis for i = 1:objects rectangle('Position',STATS(i,k).BoundingBox,... 'EdgeColor',[0 1 0]) %draw box around object Objx = STATS(i,k).Centroid(1); %centroid of object Objy = STATS(i,k).Centroid(2); %centroid of object ObjExt = floor(STATS(i,k).Extrema); if Objx <= Ix && Objy <= Iy %quadrant 2 of image text(ObjExt(5,1), ObjExt(5,2), num2str(i),... 'HorizontalAlignment', 'left','VerticalAlignment',... 'top','color',[1 1 1]); elseif Objx > Ix && Objy <= Iy %quadrant 1 of image text(ObjExt(6,1), ObjExt(6,2), num2str(i),... 'HorizontalAlignment', 'right','VerticalAlignment',... 'top','color',[1 1 1]); elseif Objx <= Ix && Objy > Iy %quadrant 3 of image text(ObjExt(2,1), ObjExt(2,2), num2str(i),... 'HorizontalAlignment', 'left','VerticalAlignment',... 'bottom','color',[1 1 1]); else %Objx > Ix && Objy > Iy %quadrant 4 of image text(ObjExt(1,1), ObjExt(1,2), num2str(i),... 'HorizontalAlignment', 'right','VerticalAlignment',... 'bottom','color',[1 1 1]); end end %save file saveas(gcf, [Treatment, ' Selection', ' Frame ', num2str(1),... '.pdf'], 'pdf'); close gcf figure,imshow(uint8(Icellsx(:,:,frames)),'InitialMagnification', 25,... 'Border', 'loose'), colorbar, title({'Frame', num2str(frames)},... 'FontSize',AxisFont) set(gca,'FontSize',10) xlabel(['pixels',' (',pixeldist,')'],'FontSize',AxisFont) ylabel(['pixels',' (',pixeldist,')'],'FontSize',AxisFont,... 'VerticalAlignment','baseline') hcfig = colorbar; %create colorbar and get handle hcTYfig = get(hcfig,'YLabel'); %get colorbar Yaxis title handle set(hcTYfig,'String',cTYfig,'Rotation',270,... 'FontSize',AxisFont,'VerticalAlignment','bottom') %insert string axis on; k = frames; %set the frame index for cell number assignment Ix = size(Icellsx,2)/2; %half length of image x-axis Iy = size(Icellsx,1)/2; %half length of image y-axis for i = 1:objects rectangle('Position',STATS(i,k).BoundingBox,... 'EdgeColor',[0 1 0]) %draw box around object Objx = STATS(i,k).Centroid(1); %centroid of object Objy = STATS(i,k).Centroid(2); %centroid of object ObjExt = floor(STATS(i,k).Extrema); if Objx <= Ix && Objy <= Iy text(ObjExt(5,1), ObjExt(5,2), num2str(i),... 'HorizontalAlignment', 'left','VerticalAlignment',... 'top','color',[1 1 1]); elseif Objx > Ix && Objy <= Iy text(ObjExt(6,1), ObjExt(6,2), num2str(i),... 'HorizontalAlignment', 'right','VerticalAlignment',... 'top','color',[1 1 1]); elseif Objx <= Ix && Objy > Iy text(ObjExt(2,1), ObjExt(2,2), num2str(i),... 'HorizontalAlignment', 'left','VerticalAlignment',... 'bottom','color',[1 1 1]); else %Objx > Ix && Objy > Iy text(ObjExt(1,1), ObjExt(1,2), num2str(i),... 'HorizontalAlignment', 'right','VerticalAlignment',... 'bottom','color',[1 1 1]); end end %save file saveas(gcf, [Treatment, ' Selection', ' Frame ', num2str(frames),... '.pdf'], 'pdf'); close gcf %clear Icellsxx %---------------------------------------------------
Plot Intensity vs Time for each cell and publish
back to top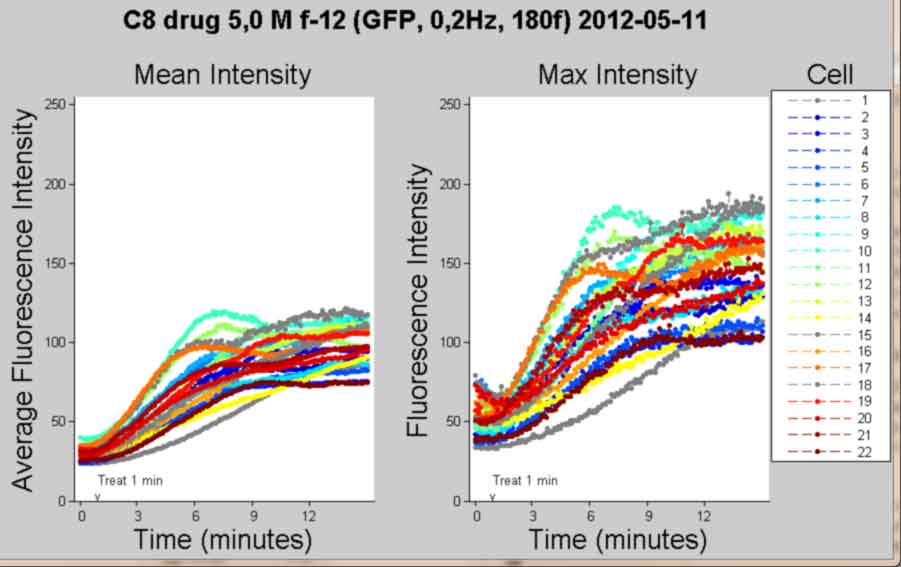
%--------------------------------------------------- PlotTitle = {[Treatment ' ' MyDate]}; % tickx = 0:60:frames; %make ticks at intervals of 60 % tickx(tickx==0) = 1; %make all zeros into ones % tickxs = cell(length(tickx),1); %preallocate % for k = 1:length(tickx) % tickxs{k} = num2str(tickx(k)); %make a cell array of tick names % end tickx = FluoXTick; %make tick intervals for markers hfig1 = figure; %handle to figure hsub1 = subplot(1,2,1); %handle to subplot hold on; %hold figure plots, dont overwrite title('Mean Intensity','FontSize',AxisFont); ylabel('Average Fluorescence Intensity','FontSize',AxisFont) if strcmp(FluoXunits,'frame') xlabel('Frame (Rate = 0.2Hz)','FontSize',AxisFont); else xlabel('Time (minutes)','FontSize',AxisFont); end X1 = (1:frames)'; if objects <= 13 && strcmp(Printcolor,'BW') for i = 1:objects Y1 = meanInt(i,1,:); Y1 = Y1(:); if ObjectOut(i) plot(X1(tickx),Y1(tickx),mymark(i),'MarkerSize',6,... 'MarkerFaceColor',[1 1 1], 'MarkerEdgeColor','k') else plot(X1(tickx),Y1(tickx),mymark(i),'MarkerSize',6,... 'MarkerFaceColor',[0 0 0], 'MarkerEdgeColor','k') end end hlegend = legend(cellnumd, 'Location',... 'NorthEastOutside'); for i = 1:objects %make diff markers for line series Y1 = meanInt(i,1,:); Y1 = Y1(:); if ObjectOut(i) plot(X1,Y1, 'color',[.5 .5 .5], 'LineStyle','-') else plot(X1,Y1, 'k-') end end else for i = 1:objects Y1 = meanInt(i,1,:); Y1 = Y1(:); if ObjectOut(i) plot(X1,Y1, '--','color',[0.5 0.5 0.5]... ,'Marker','o','MarkerSize',3,... 'MarkerFaceColor',[0.5 0.5 0.5] ) else plot(X1,Y1, '--','color',colorId(i,:)... ,'Marker','o','MarkerSize',3,... 'MarkerFaceColor',colorId(i,:) ) end end hlegend = legend(cellnumd, 'Location',... 'NorthEastOutside'); end ylim(FluoYLim); hold off; hsub2 = subplot(1,2,2); title('Max Intensity','FontSize',AxisFont); ylabel('Fluorescence Intensity','FontSize',AxisFont) if strcmp(FluoXunits,'frame') xlabel('Frame (Rate = 0.2Hz)','FontSize',AxisFont); else xlabel('Time (minutes)','FontSize',AxisFont); end hold on; if objects <= 13 && strcmp(Printcolor,'BW') for i = 1:objects Y1 = maxInt(i,1,:); Y1 = Y1(:); if ObjectOut(i) plot(X1(tickx),Y1(tickx),mymark(i),'MarkerSize',6,... 'MarkerFaceColor',[1 1 1], 'MarkerEdgeColor','k') else plot(X1(tickx),Y1(tickx),mymark(i),'MarkerSize',6,... 'MarkerFaceColor',[0 0 0], 'MarkerEdgeColor','k') end end for i = 1:objects %make diff markers for line series Y1 = maxInt(i,1,:); Y1 = Y1(:); if ObjectOut(i) plot(X1,Y1, 'color',[0.5 0.5 0.5], 'LineStyle','-') else plot(X1,Y1, 'k-') end end else for i = 1:objects Y1 = maxInt(i,1,:); Y1 = Y1(:); if ObjectOut(i) plot(X1,Y1, '--','color',[0.5 0.5 0.5]... ,'Marker','o','MarkerSize',3,... 'MarkerFaceColor',[0.5 0.5 0.5] ) else plot(X1,Y1, '--','color',colorId(i,:)... ,'Marker','o','MarkerSize',3,... 'MarkerFaceColor',colorId(i,:) ) end end end ylim(FluoYLim); htitle = text(1, 1, PlotTitle, 'FontSize', AxisFont,... 'FontWeight', 'bold', 'HorizontalAlignment',... 'center'); hold off; titlepos = [-36 303.382 17.3205]; legpos =[0.8551, 0.1112, 0.1283, 0.7641]; %position legout = [0.86, 0.1061, 0.1390, 0.7691]; %position legPBR = [1 3.1250 1]; %aspect ratio legDAR = [1 0.3200 2]; %aspect ratio sub1pos = [0.0820, 0.10, 0.32, 0.7]; %position sub1out = [0.01, -0.109, 0.42, 1.025]; %position sub2pos = [0.5, 0.10, 0.32, 0.7]; %position sub2out = [0.4492, -0.109, 0.42, 1.025]; %position set(hfig1,'PaperOrientation','landscape'); set(htitle,'Position', titlepos); set(hlegend,'FontSize', 10); set(hlegend,'Position',legpos); set(hlegend,'OuterPosition',legout); set(hlegend,'DataAspectRatio',legDAR); set(hlegend,'PlotBoxAspectRatio',legPBR); set(hsub1,'TickDir','out'); set(hsub1,'Position',sub1pos); set(hsub1,'OuterPosition',sub1out); set(hsub1,'XLim',FluoXLim); %x-axis limits set(hsub1,'XTick', FluoXTick); set(hsub1,'XTickLabel',FluoXTickS); set(hsub1,'YTick', FluoYTick); set(hsub1,'YTickLabel',FluoYTickS); set(hsub2,'TickDir','out'); set(hsub2,'Position',sub2pos); set(hsub2,'OuterPosition',sub2out); set(hsub2,'Xlim',FluoXLim); set(hsub2,'XTick', FluoXTick); set(hsub2,'XTickLabel',FluoXTickS); set(hsub2,'YTick', FluoYTick); set(hsub2,'YTickLabel',FluoYTickS); if FrameTreat ~= 0 %mark treat time if treated hsub1at = text(FrameTreat,FluoYLim(1),... '{\fontname{arial}v}',... 'Parent',hsub1,'Color',[.1 .1 .1],'Fontsize', 10,... 'HorizontalAlignment','Center',... 'VerticalAlignment','baseline'); hsub1bt = text(FrameTreat,FluoYLim(1)+10,... ['Treat ',num2str(TimeTreat), ' min'],... 'Parent',hsub1,'Color',[.1 .1 .1],'Fontsize', 10,... 'HorizontalAlignment','Left',... 'VerticalAlignment','baseline'); hsub2at = text(FrameTreat,FluoYLim(1),... '{\fontname{arial}v}',... 'Parent',hsub2,'Color',[.1 .1 .1],'Fontsize', 10,... 'HorizontalAlignment','Center',... 'VerticalAlignment','baseline'); hsub2bt = text(FrameTreat,FluoYLim(1)+10,... ['Treat ',num2str(TimeTreat), ' min'],... 'Parent',hsub2,'Color',[.1 .1 .1],'Fontsize', 10,... 'HorizontalAlignment','Left',... 'VerticalAlignment','baseline'); hlegendt = text(0.5,1,'Cell','Parent',hlegend,'units','normalized',... 'VerticalAlignment','bottom','HorizontalAlignment','center',... 'FontSize',AxisFont); end %save figure saveas(gcf, [Treatment, ' Mean and Max Intensity Plots', '.pdf'], 'pdf'); close gcf %---------------------------------------------------
Plot Intensity vs Time(frame) and publish
back to top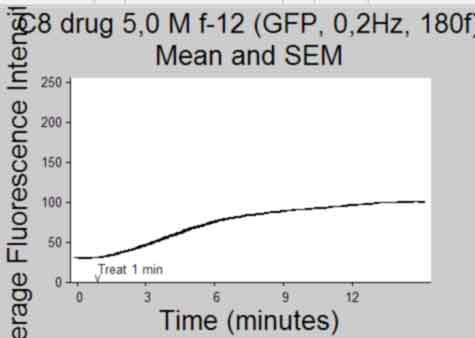
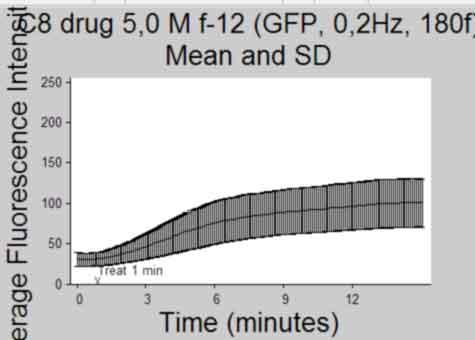
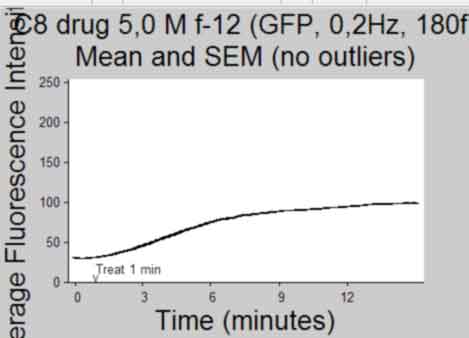
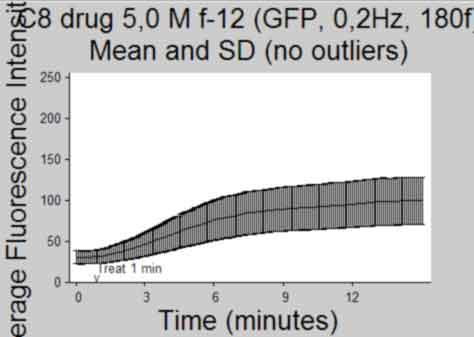
%--------------------------------------------------- Ytemp = {SemInt; StdInt; SemIntadj; StdIntadj}; Titletemp = {'Mean and SEM'; 'Mean and SD'; 'Mean and SEM (no outliers)';... 'Mean and SD (no outliers)' }; MeanIntTemp = {MeanInt; MeanInt; MeanIntadj; MeanIntadj}; for k = 1:4 if k>2 && isempty(find(ObjectOut,1)) continue %skip iteration if k>2 and if there were no outliers identified end figure; herr = errorbar(MeanIntTemp{k},Ytemp{k},'k'); %plot using errorbar function % herrleg = legend({'Mean Fluorescence Intensity'},... % 'Location','NorthEastOutside','Orientation',... % 'vertical'); title({Treatment;Titletemp{k}},'FontSize',AxisFont); %title(file) ylabel('Average Fluorescence Intensity','FontSize',AxisFont); if strcmp(FluoXunits,'frame') xlabel('Frame (Rate = 0.2Hz)','FontSize',AxisFont); else xlabel('Time (minutes)','FontSize',AxisFont); end xlim(FluoXLim) ylim(FluoYLim) haxes = gca; %get current axes handle set(haxes,'TickDir','out', 'box','off'); set(haxes,'XTick', FluoXTick); %set xticks set(haxes,'XTickLabel', FluoXTickS,'FontSize',10); %set the Xlabels set(haxes,'YTick', FluoYTick); %set yticks set(haxes,'YTickLabel', FluoYTickS,'FontSize',10); %set the Ylabels if FrameTreat ~= 0 %mark treat time if treated h3atxt = text(FrameTreat,FluoYLim(1),... '{\fontname{arial}v}',... 'Color',[.1 .1 .1],'Fontsize', 10,... 'HorizontalAlignment','Center',... 'VerticalAlignment','baseline'); h3btxt = text(FrameTreat,FluoYLim(1)+10,... ['Treat ',num2str(TimeTreat), ' min'],... 'Color',[.1 .1 .1],'Fontsize', 10,... 'HorizontalAlignment','Left',... 'VerticalAlignment','baseline'); end %save figure saveas(gcf, [Treatment, ' ', Titletemp{k}, ' Plot', '.pdf'], 'pdf'); close gcf end clear Ytemp Titletemp MeanIntTemp %-----------------------------------------------
Plot a Curve Fit to response after treatment
back to top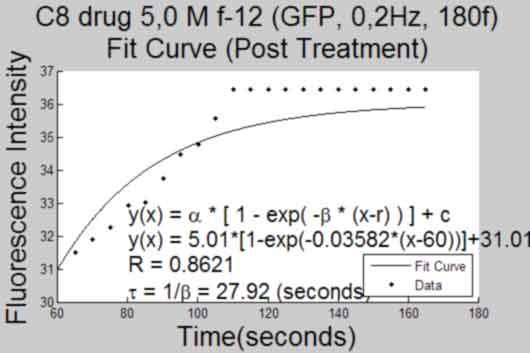
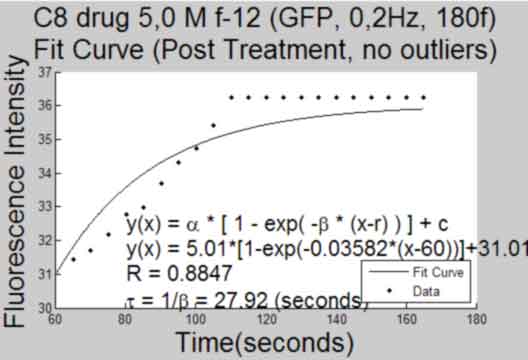
%----------------------------------------------- if FrameTreat ~= 0 fitcurve = []; goodness = []; TitleTemp = 'Fit Curve (Post Treatment)'; eval('[fitcurve goodness] = myfit2(MeanInt,FrameTreat)') %get fit curve to data with outliers set(gca,'Box','off'); gcf; title({Treatment; TitleTemp},'FontSize',AxisFont); saveas(gcf,[Treatment, ' ', TitleTemp, 'Plot', '.pdf'], 'pdf'); close gcf; if ~isempty(find(ObjectOut,1)) fitcurveadj = []; goodnessadj = []; TitleTemp = 'Fit Curve (Post Treatment, no outliers)'; eval('[fitcurveadj goodnessadj] = myfit2(MeanIntadj,FrameTreat)') %get fit curve to data without outliers set(gca,'Box','off'); gcf; title({Treatment; TitleTemp},'FontSize',AxisFont); saveas(gcf,[Treatment, ' ', TitleTemp, 'Plot', '.pdf'], 'pdf'); close gcf; end end %-----------------------------------------------
Save Variables
back to top%----------------------------------------------- if FrameTreat ~= 0 && ~isempty(find(ObjectOut,1)) %save when Treated and when outliers exist save([Treatment,' variables','.mat'], 'MeanInt',... 'STATS','SemInt','StdInt','maxInt','meanInt','semInt','fitcurve',... 'goodness','fitcurveadj','goodnessadj'); elseif FrameTreat ~= 0 && isempty(find(ObjectOut,1)) %save when Treated and no outliers save([Treatment,' variables','.mat'], 'MeanInt',... 'STATS','SemInt','StdInt','maxInt','meanInt','semInt','fitcurve',... 'goodness'); elseif FrameTreat == 0 && ~isempty(find(ObjectOut,1)) %save when not treated and outliers exist save([Treatment,' variables','.mat'], 'MeanInt',... 'STATS','SemInt','StdInt','maxInt','meanInt','semInt'); else %save when not treated and no outliers save([Treatment,' variables','.mat'], 'MeanInt',... 'STATS','SemInt','StdInt','maxInt','meanInt','semInt'); end %-----------------------------------------------
Plot Radialwise Intensity and publish
back to top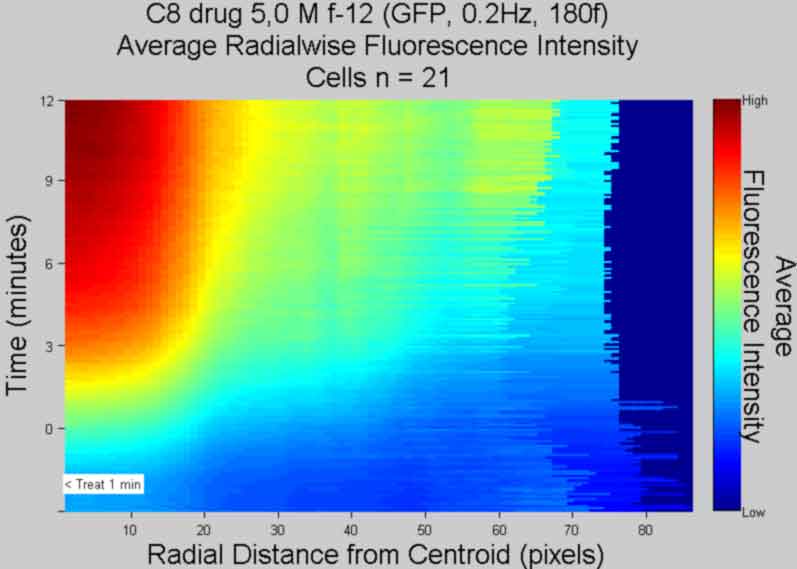
%----------------------------------------------- %%Create Rings % rings will overlap betwen adjacent radii because % of discrete shape of image pixels %%Set Constant Parameter & preallocate arrays Theta = linspace(0,2*pi,40); Ring = cell(objects,frames); %create cell to store cell r data cellboxw = 1:objects; cellboxh = cellboxw; Cellsub = cell(objects,1); %create cell array to store cropped cells %%Get Parameters one frame at a time for fr = 1:frames; %iterate through frames for i = 1:objects %iterate through objects centroid1 = floor(STATS(i,fr).Centroid); uLcorner1 = ceil(STATS(i,fr).BoundingBox(1:2)); centroid1sub = centroid1 - uLcorner1 + 1; %get sub centroid coordinates cellboxw(i) = ceil(STATS(i,fr).BoundingBox(3))-1; cellboxh(i) = ceil(STATS(i,fr).BoundingBox(4))-1; cellboxW = uLcorner1(1):uLcorner1(1)+cellboxw(i);%turn over for each object cellboxH = uLcorner1(2):uLcorner1(2)+cellboxh(i); %turn over for each object Cellsub{i,1} = Icellsx(cellboxH,cellboxW,fr); %turn over for ea frame rlim = ceil(1.415*max([cellboxw(i),cellboxh(i)])/2); %find max angular radii for object i Ring{i,fr} = cell(rlim,1); %create cell arrays in Ring to store radii data %Make circle r1 for k=1 %iterate through r1 r = k; %radius of circle in pixels Rho = ones(1,40)*r; %create array of angles [X Y] = pol2cart(Theta,Rho); %convert to cartesian X = ((X+centroid1sub(1))); %shift center of circles Y = (Y+size(Cellsub{i,1},1)-centroid1sub(2)+1); BWr = roipoly(Cellsub{i,1},X,Y); %use Cellsub vs Template %BWr = roipoly(Template,X,Y); %create bw template for r BWr = bwmorph(BWr,'spur'); %remove "thorns" [R C] = find(BWr); %find all coordinates of ring Idx = sub2ind(size(BWr),R,C); %convert coordinates to linear index Ring{i,fr}{k,1}(1,:) = Idx; %store linear index V = Cellsub{i,1}(Idx); %get intensity Ring{i,fr}{k,1}(2,:) = V; %store intensity clear C R X Y V end %Make circles 2-rlim %For loop start with ring 3 end with ring rlim %counter will be set to k, create r=k+2 and r=k rings for k = 2:rlim %iterate through r2+ r = k; %radius of circle in pixels, not inclusive Rho = ones(1,40)*r; %create array of angles [X Y] = (pol2cart(Theta,Rho)); %convert to cartesian X = ((X+centroid1sub(1))); %shift center of circles Y = (Y+size(Cellsub{i,1},1)-centroid1sub(2)+1); BWr = roipoly(Cellsub{i,1},X,Y); %create bw template for r BWr = bwmorph(BWr,'spur'); BWr = bwmorph(BWr,'remove'); clear X Y [R C] = find(BWr); %get coordinates of ring if isempty(R) %stop iterating through radii if radii rings fall off break end Idx = sub2ind(size(BWr),R,C); %convert coordinates to linear index V = Cellsub{i,1}(Idx); %get intensity if r >= round(1/1.415*(rlim)) %start checking values at orthoganol radii if sum(V) == 0 %stop writing values if equal to zero break end end Ring{i,fr}{k,1}(1,:) = Idx; %store index Ring{i,fr}{k,1}(2,:) = V; %store intensity clear C R X Y V end end end clear BWr* cellbox* Cellsub %%Go through all frames and compute the average r values for each object %%and for all cells %Robjects = cell(objects,1); Rframes = cell(1,frames); %replace second entry with frames for i = 1:frames %iterate through frames for j = 1:size(Ring(:,i),1) %iterate through objects a(j) = size(Ring{j,i},1); %get size of radii for each object in frame end a = max(a); %get max radii size for frame i Rframes{1,i} = cell(a,1); %preallocate cell size of length max radii for j = 1:objects %iterate through objects for k = 1:size(Ring{j,i},1) %iterate through radii %Store radii collapse objects, maintain frames Rframes{1,i}{k,1} = cat(2,Rframes{1,i}{k,1},... Ring{j,i}{k,1}(2,:)); end end end clear Ring %%Find the mean value for radii of each frame for i = 1:frames %iterate through frames for j = 1:size(Rframes{1,i},1) %iterate through radii Rmeanframes{1,i}(j,1) = mean(nonzeros(Rframes{1,i}{j,1})); Rstd = std(nonzeros(Rframes{1,i}{j,1})); Rstderrframes{1,i}(j,1) = Rstd/sqrt(nnz(Rframes{1,i}{j,1})); end end clear Rframes %create a matrix of radii %Yaxis Frame # %Xaxis Radial distance from centroid %Zaxis Average Radialwise Fluorescence Intensity %A = cell2mat(Rmeanframes); %create a matrix, might error if dim wrong A = []; for k = 1:frames %iterate through frames A(1:size(Rmeanframes{1,k},1),k) = Rmeanframes{1,k}(:); %(RFIxFrame) end A = fliplr(A); %flip matrix columns, mirror the frame axis (FramexRFI) hfsurf = figure; %create figure and get handle hsurf = surf(A); %create mesh plot for Rmeanframes if strcmp(Printcolor,'BW') colormap gray else colormap jet end title({Treatment;... 'Average Radialwise Fluorescence Intensity';... ['Cells n = ', num2str(objects)] },'FontSize',AxisFont) %update if strcmp(FluoXunits,'minutes') xlabel('Time (minutes)','FontSize',AxisFont) else xlabel('Frame (Rate = 0.2Hz)','FontSize',AxisFont) end ylabel('Radial Distance from Centroid (pixels)','FontSize',AxisFont) zlabel('Radialwise Average Fluorescence Intensity','FontSize',AxisFont) set(hsurf,'EdgeColor','flat') %set edge box color set(hfsurf,'Position',[50 250 550 400]); %position figure window view(90,90); %change surf orientation hcsurf = colorbar; %create colorbar and get handle % xtickdiv = frames/6; %create x-axis divisions space % xtickdiv = 0:xtickdiv:frames; %create x-axis % xtickdiv(xtickdiv==0)=1; %make 0 = 1 haxes = gca; %get current axes handle set(haxes,'XLim',[1 frames]) %set x-axis limits set(haxes,'XTick',FluoXTick) %set x-axis ticks set(haxes,'XTickLabel',flipud(FluoXTickS)) %write reverse labels % xtickL = get(haxes,'XTick'); %get x-axis labels % xtickL = fliplr(xtickL); %make the reverse of x-axis labels % set(haxes,'XTickLabel', xtickL); %write reverse the Xlabels radialLim = size(A); set(haxes,'Ylim',[1 radialLim(1)]) set(haxes,'XGrid','off','YGrid','off','ZGrid','off') set(haxes,'TickDir','out') cTLsurf = get(hcsurf,'YLim'); %get colorbar limits set(hcsurf,'YTick',cTLsurf) %create ticks at limits set(hcsurf,'YTickLabel',{'Low','High'}) %create Yticklabels cTYSsurf = {'Average'; 'Fluorescence Intensity'}; %create colorbar title string hcTYsurf = get(hcsurf,'YLabel'); %get colorbar Yaxis title handle set(hcTYsurf,'String',cTYSsurf,'Rotation',270,... 'VerticalAlignment','baseline','FontSize',AxisFont) %insert string %tightmap %eliminate/reduce white boarder ztemp = zlim; ztemp = ztemp(2); if FrameTreat ~= 0 %mark treat time if treated text(frames-FrameTreat,1,ztemp,['< Treat ',num2str(TimeTreat),' min'],... 'fontsize',10,'Color',[.1 .1 .1],... 'BackgroundColor','w') end %save file saveas(gcf, [Treatment, ' Average Radial Fluorescence', '.pdf'], 'pdf'); close gcf %-----------------------------------------------
Save Variables
back to top%----------------------------------------------- if FrameTreat ~= 0 save([Treatment,' variables','.mat'], 'Treatment','MeanInt','Rmeanframes','Rstderrframes',... 'STATS','SemInt','StdInt','maxInt','meanInt','semInt'); else save([Treatment,' variables','.mat'], 'Treatment','MeanInt','Rmeanframes','Rstderrframes',... 'STATS','SemInt','StdInt','maxInt','meanInt','semInt','fitcurve','goodness'); end %-----------------------------------------------
Make Avi Movie of original images and Mean +/- SD Inensity of Cell Selection
back to top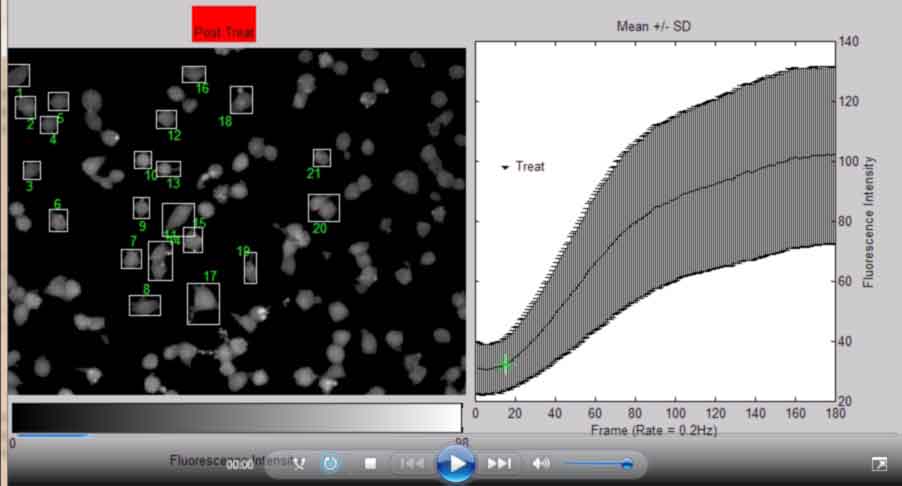
%----------------------------------------------- %Open video object and append frames from Image stack aviobj = VideoWriter([Treatment,' ', MyDate,'.avi']); %make file name aviobj.FrameRate = 12; %frames per second aviobj.Quality = 100; %percent quality open(aviobj); %open an object file for k = 1:frames; fig = figure(1); h12=subplot(1,2,1); imshow(Icellsx(:,:,k),'DisplayRange',... [min(min(Icellsx(:,:,k))),max(max(Icellsx(:,:,k)))],... 'InitialMagnification', 25,'Border', 'loose'); %do not scale 8bit map %imshow(Icellsx(:,:,k)),... hcb = colorbar('location','SouthOutside'); if k < FramePostTreat; title({'Pre Treat'; []},'backgroundcolor',[0 1 0]); else title({[]; 'Post Treat'},'backgroundcolor',[1 0 0]); end htxt = text(1,1.35,{['NSC-34 ', Passage, ' ', Treatment,' ', MyDate,];... ['Frame ', num2str(k),' of ', num2str(frames),... FrameRate]}, 'Fontsize', 18,'HorizontalAlignment',... 'Center', 'Units','normalized'); h22=subplot(1,2,2); errorbar(MeanInt,StdInt,['k','-']), hold on,... %hleg = legend('mean', 'max','Location', 'SouthOutside'); plot(k,MeanInt(k),['b','*']),... %plot(k,maxall(k), ['k','*']),... title('Mean +/- SD'), ylabel('Fluorescence Intensity'),... xlabel('Frame (Rate = 0.2Hz)'), xlim([0 frames]); h2txt = text(12,ceil(0.95*(max(max(MeanInt)))),... {['{\fontname{Webdings}6} ','Treat']},... 'Fontsize', 10, 'HorizontalAlignment','Left'); hold off; axis(h12,'image'),axis(h12,'off'); set(h12,'Position',[0 0 0.6 1]); set(h12,'OuterPosition',[-0.08 -0.08 0.6 1]); axis(h22,'square'); set(h22,'OuterPosition',[0.49 -0.4 0.5 1.5]); set(h22,'Position',[0.52 -0.07 0.4 1.07]); set(h22,'YAxisLocation', 'right'); %set(hleg,'Position',[0.67 0.05 0.16 0.1]) set(hcb,'XTick',[0 max(max(Icellsx(:,:,k)))]); set(hcb,'Position',[0.005 0.1 0.5 0.0500]); xlabel(hcb,'Fluorescence Intensity'); % for i = 1:objects %original bounding box drawing % if k < FramePostTreat % rectangle('Position',STATS(i,k).BoundingBox,... % 'EdgeColor',[0 0 1],'Parent',h12) % else % rectangle('Position',STATS(i,k).BoundingBox,... % 'EdgeColor',[0 0 1],'Parent',h12) % end % end for i = 1:objects %new bounding box drawing with numbers rectangle('Position',STATS(i,k).BoundingBox,... 'EdgeColor',[0 0 1],'Parent',h12) %draw box around object Objx = STATS(i,k).Centroid(1); %centroid of object Objy = STATS(i,k).Centroid(2); %centroid of object ObjExt = floor(STATS(i,k).Extrema); if Objx <= Ix && Objy <= Iy %quadrant 2 of image text(ObjExt(5,1), ObjExt(5,2), num2str(i),... 'HorizontalAlignment', 'left','VerticalAlignment',... 'top','color',[1 1 1],'Parent',h12); elseif Objx > Ix && Objy <= Iy %quadrant 1 of image text(ObjExt(6,1), ObjExt(6,2), num2str(i),... 'HorizontalAlignment', 'right','VerticalAlignment',... 'top','color',[1 1 1],'Parent',h12); elseif Objx <= Ix && Objy > Iy %quadrant 3 of image text(ObjExt(2,1), ObjExt(2,2), num2str(i),... 'HorizontalAlignment', 'left','VerticalAlignment',... 'bottom','color',[1 1 1],'Parent',h12); else %Objx > Ix && Objy > Iy %quadrant 4 of image text(ObjExt(1,1), ObjExt(1,2), num2str(i),... 'HorizontalAlignment', 'right','VerticalAlignment',... 'bottom','color',[1 1 1],'Parent',h12); end end F = getframe(gcf); %get current figure handle writeVideo(aviobj,F); %write current frame to aviobj end close(gcf); close(aviobj);
back to top