Contents
To do
back to top
Initialize variables
back to top
clear all;
close all;
clc;
h = waitbar(0,'Initializing Global waitbar...',...
'position',[400 100 275 50]);
button = [];
button1 = [];
button2 = [];
button3 = [];
button4 = [];
button5 = [];
button6 = [];
directory = dir('*abf');
fig = 0;
step = 0;
Load data
back to top
for n = 1:size(directory, 1);
x = abfload1(directory(n).name);
x0 = [];
x1 = [];
marks = [];
hfcount = 0;
for k = 1:size(x,3);
x0(:,1,k) = x(1:700, 1, k);
x1(:,1,k) = x0(:,1,k) - mean(x0(1:20,1,k));
x2 = x1(:,:,k);
if k == 1
[M1, M2, M3] = markers(x2);
marks(:,1,k) = M1;
marks(:,2,k) = M2;
marks(:,3,k) = M3;
elseif var(x2(1:200)) <= 0.01
marks(:,1,k) = marks(:,1,k-1);
marks(:,2,k) = marks(:,2,k-1);
marks(:,3,k) = marks(:,3,k-1);
if k < 46
hfcount = hfcount+1;
hf(hfcount) = k;
end
else
[M1, M2, M3] = markers(x2);
marks(:,1,k) = M1;
marks(:,2,k) = M2;
marks(:,3,k) = M3;
end
end
time = linspace(0,7,700)';
correct = 0;
while correct ~= 1
button = questdlg({['Would you like to be able to edit the ',...
'placement of popspike markers for '];...
['Mouse #', strtok((directory(n).name), '.')]},...
'Analysis Option', 'Yes', 'No', 'No');
button = lower(button);
if strcmp(button, 'yes') || strcmp(button, 'no')
correct = 1;
else correct =0;
end
end
Display Population Spike Automation
back to top
if strcmp(button, 'yes')
if isempty(button1)
button1 = questdlg({'Watch placement of Markers';...
['Keep in mind or record the waveform #(s) ',...
'that appear incorrect'];...
['You will later have the opportunity to ', ...
'modify the waveform(s) that need edits']},...
'Reminder', 'Ok', 'Ok');
end
hh = waitbar(0,'Initializing Current progress waitbar...',...
'position',[400 200 275 50]);
for i = 1:size(x1,3)
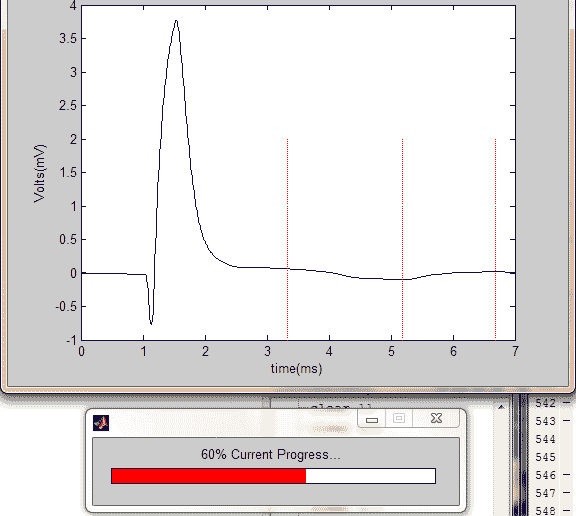
pic = figure(1);
wave = x1(:,:,i);
plot(time',wave); hold on;
plot(time(marks(2,1,i)), linspace(-1,2), 'r',...
time(marks(2,2,i)), linspace(-1,2),...
'r', time(marks(2,3,i)), linspace(-1,2), 'r');
title({['Mouse #', strtok((directory(n).name), '.')];...
['Waveform #', int2str(i)]});
xlabel('time(ms)');
ylabel('Volts(mV)');
hold off;
pause(.2);
perc = i/size(x1,3);
waitbar(perc,hh,sprintf('%.0f%% Current Progress...',perc*100))
end
close(pic);
close(hh);
Prompt User to Edit Automated PS
back to top
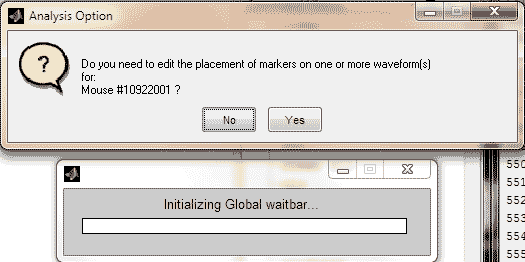
button2 = questdlg({['Do you need to edit the placement ',...
'of markers on one or more waveform(s) for:'];
['Mouse #', strtok((directory(n).name), '.'), ' ?']},...
'Analysis Option', 'No', 'Yes', 'No');
button2 = lower(button2);
button5 = [];
while ~strcmp(button5,'no')
if strcmp(button2, 'yes')
rangetest = 0;
while rangetest == 0
prompt = ({['Enter comma separated Range of ',...
'waveform #(s) that need editing Example: 12,19']});
dlg_title = 'Enter comma separated Range of Waveform #(s)';
num_lines = 1;
options.Resize='on';
options.WindowStyle='normal';
options.Interpreter='tex';
def = {'1,1'};
Range = inputdlg(prompt,dlg_title,num_lines,def,options);
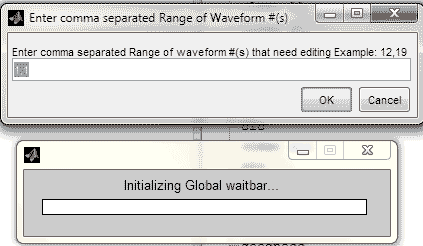
range = Range{1};
range = str2num(range);
if length(range) < 2 || length(range) > 2
rangeh = warndlg({'Enter comma separated range';...
'For Example:'; '12,19'});
uiwait(rangeh);
continue
elseif range(1) == 0 || range(2) > size(x1,3)
rangeh = warndlg(['Range should be from 1 to ',...
num2str(size(x1,3))]);
uiwait(rangeh);
continue
elseif range(1) > range(2)
rangeh = warndlg('For Range [a,b], b must be > a');
uiwait(rangeh);
else
rangetest = 1;
end
end
end
if strcmp(button2, 'yes')
hh = waitbar(0,'Initializing Current progress waitbar...',...
'position',[400 200 275 50]);
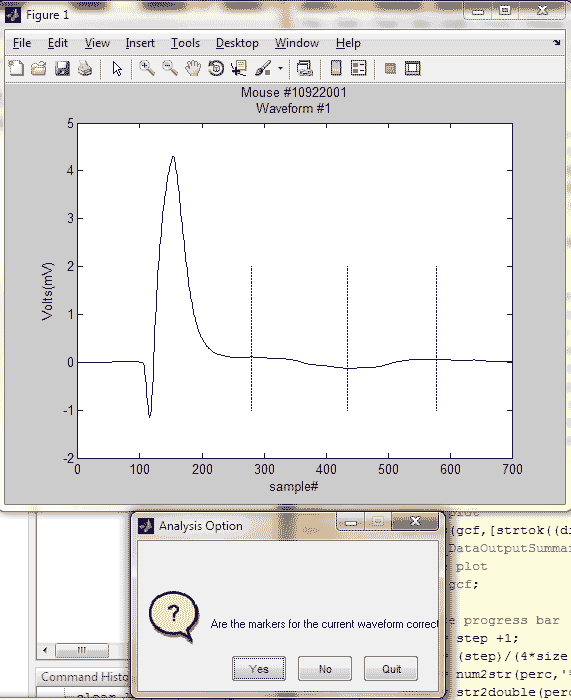
for i = range(1):range(2)
pic = figure(1);
wave = x1(:,:,i);
plot(1:length(wave),wave); hold on;
plot(marks(2,1,i), linspace(-1,2), 'b',...
marks(2,2,i), linspace(-1,2),...
'b', marks(2,3,i), linspace(-1,2), 'b');
title({['Mouse #', strtok((directory(n).name), '.')];...
['Waveform #', int2str(i)]});
xlabel('sample#');
ylabel('Volts(mV)');
button3 = myquestdlg(['Are the markers for the ',...
'current waveform correct?'], 'Analysis Option',...
'Yes', 'No','Quit','Yes');
button3 = lower(button3);
if strcmp(button3, 'quit')
hold off;
break;
end
if strcmp(button3, 'no')
yx = [];
if i == range(1) && isempty(button4);
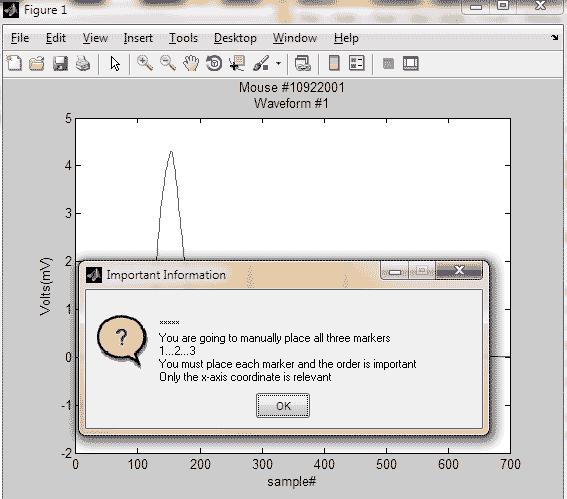
button4 = questdlg({'*****';...
['You are going to manually place ',...
'all three markers'];...
'1...2...3'; ['You must place each marker',...
' and the order is important']; ...
'Only the x-axis coordinate is relevant'},...
'Important Information', 'OK', 'OK');
end
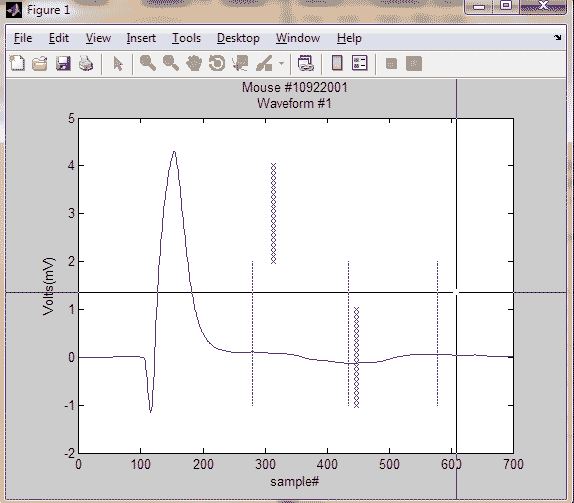
but = 0;
while but ~= 3
[xi,yi,click] = ginput(1);
if isempty(click)
return;
elseif click ~= 1
continue;
elseif ceil(xi+1) > length(wave)
rangeh = warndlg(['Stay in bounds',...
'& reselect preveious marker']);
uiwait(rangeh);
continue;
else
but = but+1;
end
xi = ceil(xi);
yi = ceil(yi);
plot(xi,yi-1:.1:yi+1,'rx')
yx(:,but) = [wave(xi+1);xi];
end
hold off;
marks(:,1,i) = yx(:,1);
marks(:,2,i) = yx(:,2);
marks(:,3,i) = yx(:,3);
end
hold off;
close(pic);
perc = (i-range(1)+1)/(range(2)-range(1)+1);
waitbar(perc,hh,sprintf('%.0f%% Current Progress...'...
,perc*100))
end
close(hh);
end
button5 = questdlg({['Do you want to continue editing ',...
'the placement of markers for:'];...
['Mouse #', strtok((directory(n).name), '.')]},...
'Analysis Option', 'No', 'Yes', 'No');
button5 = lower(button5);
end
end
Consolidate PS Data Collection
back to top
POP = [];
if size(x,3) >= hf(length(hf)) + 100
for k = 1:hf(length(hf)) + 100;
POP(k) = (1/2*(marks(1,1,k) + marks(1,3,k))) - marks(1,2,k);
end
else
for k = 1:size(x,3);
POP(k) = (1/2*(marks(1,1,k) + marks(1,3,k))) - marks(1,2,k);
end
end
for k = 1:length(hf);
POP(hf(k)) = 0;
end
POPave = mean(POP(1:32));
POPN = 100*(POP/POPave);
POPNave = mean(POPN(112:121));
if length(POPN) >= 136;
POPN_1 = POPN((hf(length(hf)) + 1):136);
else
POPN_1 = POPN((hf(length(hf)) + 1):length(POPN));
end
Y = POPN_1;
Y = Y - min(Y);
fitlimit = length(Y);
k = 0:1/3:length(POPN)/3;
if length(k) ~= length(POPN)
TIME = k(1:length(POPN));
end
if length(k) ~= length(Y)
k = k(1:length(Y));
end
Plot POPN vs TIME & Yfit vs Time & Summar Page
back to top
figure(fig+n);
set(gcf,'name',[strtok((directory(n).name), '.'), ' ',...
'POPN vs time(min)']);
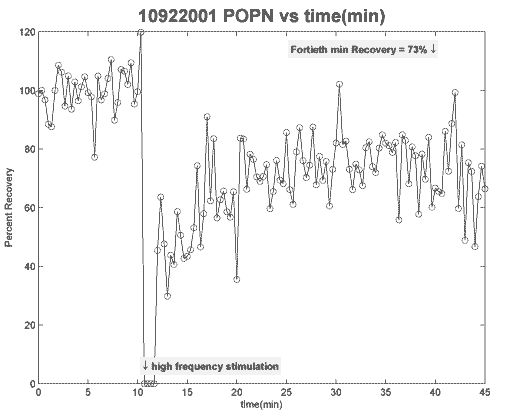
plot(TIME, POPN,['b','o']); hold on; plot(TIME, POPN,['k','-']);
title([strtok((directory(n).name), '.'), ' ',...
'POPN vs time(min)'], 'fontsize', 18, 'fontweight', 'bold');
xlabel('time(min)');
ylabel('Percent Recovery');
text(ceil(hf(1)*1/3)-.5,.05*diff(ylim)+min(ylim),...
'\downarrow high frequency stimulation',...
'fontweight', 'bold', 'horizontalalignment', 'left',...
'backgroundcolor', 'y');
text(40,.95*diff(ylim)+min(ylim),['Fortieth min Recovery = '...
,num2str(POPNave,'%2.0f'),'% \downarrow'], 'fontweight',...
'bold', 'horizontalalignment', 'right', 'backgroundcolor', 'y');
hold off;
fig = fig + 1;
saveas(gcf,[strtok((directory(n).name), '.'), '_POPNvstime'],'pdf');
close gcf;
step = step +1;
perc = (step)/(4*size(directory,1));
perc = num2str(perc,'%0.2f');
perc = str2double(perc);
waitbar(perc,h,sprintf('%.0f%% Global Progress...',perc*100))
fitlimit = length(Y);
button4 = [];
while strcmp(button4, 'no') ~= 1
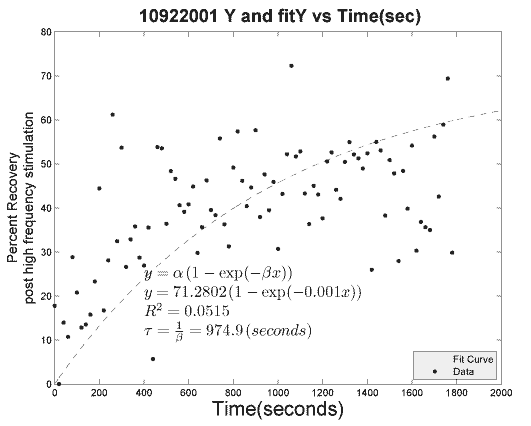
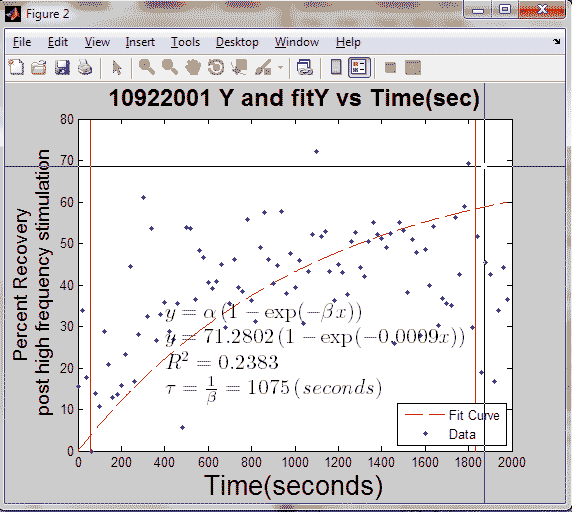
[curvefit,goodness,fitx] = myfitY(Y,'yes',20,'%PS',fig+n);
Tao = 1/curvefit.beta;
R = abs(goodness.rsquare);
dfe = goodness.dfe;
rmse = goodness.rmse;
sse = goodness.sse;
set(gca,'nextplot','replacechildren');
title([strtok((directory(n).name), '.'), ' ',...
'Y and fitY vs Time(sec)'], 'fontsize', 18, 'fontweight', 'bold');
ylabel({'Percent Recovery';...
'post high frequency stimulation'}, 'fontsize', 14);
button4 = myquestdlg(['Do you need to edit the ',...
'limit of the curve fit?'],...
'Analysis Option', 'No', 'Yes', 'No');
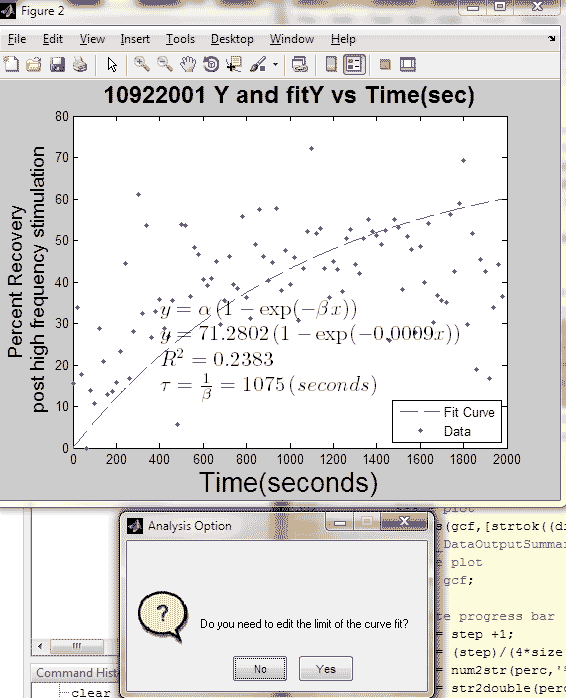
button4 = lower(button4);
if strcmp(button4, 'yes')
XCropp = [];
click_count = 0;
lastpoint = 1;
pointsY = axis;
pointsY = pointsY(3:4);
while lastpoint == 1
[xi,yi,lastpoint] = ginput(1);
line([xi,xi],pointsY,'color','r')
click_count = click_count+1;
XCropp(:,click_count) = xi;
end
startx = round(min(XCropp(:))/20);
if startx >= 0.5*length(Y);
startx = 1;
end
endx = round(max(XCropp(:))/20);
if endx <= 0.5*length(Y);
endx = length(Y);
end
Y = Y(startx:endx);
limitsCrop = [startx,endx];
hold off;
end
fitY = curvefit.alpha*(1 - exp(-curvefit.beta *(fitx)));
end
fig = fig + 1;
saveas(gcf,[strtok((directory(n).name), '.'),...
'_YandfitYvsTime'],'pdf');
close gcf;
step = step +1;
perc = (step)/(4*size(directory,1));
perc = num2str(perc,'%0.2f');
perc = str2double(perc);
waitbar(perc,h,sprintf('%.0f%% Global Progress...',perc*100))
figure(fig+n);
set(gcf,'name',[strtok((directory(n).name), '.'), ' ',...
'Output Data']);
axis([0,20,0,20]);
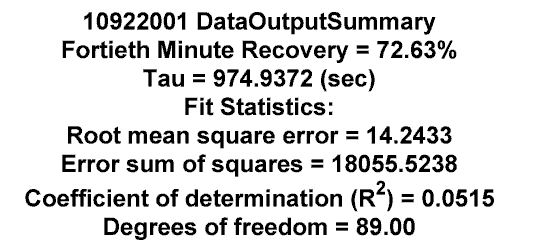
text(9.54,10,{[strtok((directory(n).name), '.'),...
' DataOutputSummary'];...
['Fortieth Minute Recovery = ',num2str(POPNave, '%2.2f'),'%'];...
['Tau = ', num2str(Tao,'%2.4f'), ' (sec)']; 'Fit Statistics:';...
['Root mean square error = ',num2str(rmse,'%2.4f')];...
['Error sum of squares = ',num2str((sse),'%2.4f')];...
['Coefficient of determination (R^2) = ', num2str(R,'%2.4f')];...
['Degrees of freedom = ', num2str(dfe,'%2.2f')]},...
'horizontalalignment', 'center','fontweight', 'bold',...
'backgroundcolor', 'w', 'verticalalignment', 'middle',...
'fontsize', 24);
axis off;
saveas(gcf,[strtok((directory(n).name), '.'),...
'_DataOutputSummary'],'pdf');
close gcf;
step = step +1;
perc = (step)/(4*size(directory,1));
perc = num2str(perc,'%0.2f');
perc = str2double(perc);
waitbar(perc,h,sprintf('%.0f%% Global Progress...',perc*100))
Pack Data into text file for export
back to top
stArray = struct;
stArray(n).name = ['mouse_', strtok((directory(n).name), '.')];
stArray(n).waveform = x1;
stArray(n).markers = marks;
stArray(n).POPN = POPN;
stArray(n).POPNave = POPNave;
stArray(n).Y = Y;
stArray(n).fitY = fitY;
stArray(n).Time = TIME;
w=size(stArray(n).waveform,3);
l=size(stArray(n).Y,2);
q=length(stArray(n).POPN);
stWrite = struct;
stWrite(n).name = stArray(n).name;
stWrite(n).waveform = reshape(stArray(n).waveform,[700,w]);
stWrite(n).markers = reshape(stArray(n).markers(1,:,:),[w,3]);
stWrite(n).POPN = reshape(stArray(n).POPN,[q,1]);
stWrite(n).POPNave = stArray(n).POPNave;
stWrite(n).Y = reshape(stArray(n).Y,[l,1]);
stWrite(n).fitY = reshape(stArray(n).fitY,[l,1]);
stWrite(n).Time = reshape(stArray(n).Time,[q,1]);
A = zeros(700,size(x0,3)+11);
A(1:length(stWrite(n).Time),1) = stWrite(n).Time;
A(1:length(time),2) = time;
A(1:length(stWrite(n).markers),3) = stWrite(n).markers(:,1);
A(1:length(stWrite(n).markers),4) = stWrite(n).markers(:,2);
A(1:length(stWrite(n).markers),5) = stWrite(n).markers(:,3);
A(1:length(stWrite(n).POPN),6) = stWrite(n).POPN;
A(1,7) = stWrite(n).POPNave;
A(1,8) = Tao;
A(1,9) = R;
A(1:length(stWrite(n).Y),10) = stWrite(n).Y;
A(1:length(stWrite(n).fitY),11) = stWrite(n).fitY;
for j = 12:size(x0,3)+11
A(:,j) = stWrite(n).waveform(:,j-11);
end
head = [];
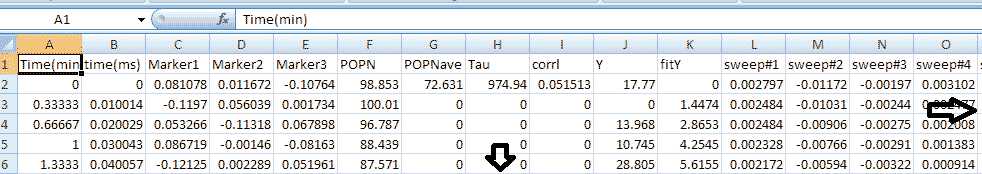
head1 = ['Time(min),','time(ms),','Marker1,','Marker2,',...
'Marker3,','POPN,','POPNave,','Tau,','corrl,','Y,','fitY,'];
head2 = 'sweep#1,';
for j = 2:size(x0,3)
head2 = [head2,'sweep#', num2str(j), ','];
end
head = [head1, head2];
format1 = '%s ';
for j = 1:size(x0,3)+10
format1 = [format1, '%s '];
end
format1 = [format1, '%s'];
fid = fopen([strtok((directory(n).name), '.'),...
'_OutputData', '.txt'], 'wt');
fprintf(fid, format1, head);
fclose(fid);
dlmwrite([strtok((directory(n).name), '.'), '_OutputData', '.txt'],...
A, '-append', 'delimiter', ',', 'roffset',1);
if n==1
fids = fopen('Summary of Output Data.txt', 'wt');
fprintf(fids, '%s %s %s', ['subjectID#,','%Recovery,','Tau']);
end
fprintf(fids, '\n%s %s %s', [[strtok((directory(n).name), '.'),','],...
[num2str(POPNave, '%2.2f'),','], [num2str(Tao,'%2.2f'),',']]);
step = step +1;
perc = (step)/(4*size(directory,1));
perc = num2str(perc,'%0.2f');
perc = str2double(perc);
waitbar(perc,h,sprintf('%.0f%% Global Progress...',perc*100))
if n ~= size(directory,1)
button6 = myquestdlg(['Do you want to stop ',...
'analyzing the data?'],...
'Analysis Option', 'No', 'Yes', 'No');
button6 = lower(button6);
if strcmp(button6, 'yes')
break;
end
end
end
fclose(fids);
close(h);
Data Files Generated
back to top